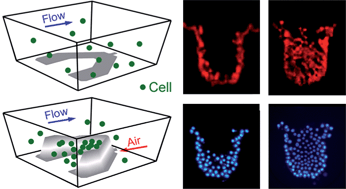
一种整合气动微结构的微流控芯片用于动态捕获细胞及高通量细胞图案化。
Introduction
微流控捕获系统在微米分辨率上操控细胞的空间排列和移动上具有多功能,且可以减少样本和试剂消耗。已经有不同的被动的细胞捕获操纵微流控芯片,细胞位于生物分子/化学固定的微区域或者禁止的固体支撑(如微孔和微坝)。其优势在于:简便的操作、容易捕获而不需要辅助设备和出色的通量。然而这些被动方法通常不可逆(如只能捕获一次或不能释放),因此可以更动态、实时和精确操作细胞捕获的积极捕获策略是十分必要的。
已经提出很多出众和有效的使用外部间接力量的动态细胞捕获方法,如介电泳、磁力、光学和声波力。但是这些方法以其需要复杂的准备、昂贵的设备及有可能对活的生物样本造成潜在伤害而受限。外部直接力量或简单的机械驱动策略也被提出,通过诱导微流控元件的尺寸变化,从而有效地控制流体/微物体的流动。 例如,压力驱动阀的机械运动已被用于分离微球,以制备原位亲和柱和反馈式计数自动化。然而同时具备可操纵性(可逆的捕获)和动态定量(即可改变的细胞捕获数)的细胞捕获仍有待探索。
此研究证明了双动态的捕获细胞可以通过可调节的气动微结构(PμS)简单、便利地实现。此外通过特别设计的PμS柱可以实现芯片上不同数目的高通量的细胞图案化。
Fig. 1 Pneumatic microstructure
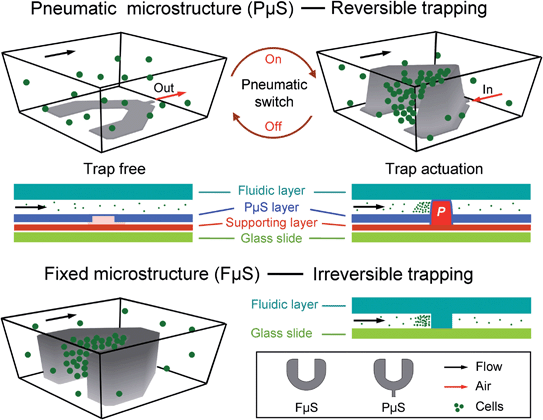
气动微结构(PμS)显示了高度可调的几何特性,因此与固定的微结构相比(FμS),可以进行可逆的细胞捕获。三维和截面图显示了用于微流控细胞捕获的经典U型设计,并将其用于本研究中的PμS辅助的细胞捕获研究。
Fig. 2 Microfluidic cell-trapping device integrated with pneumatic microstructure (PμS) arrays
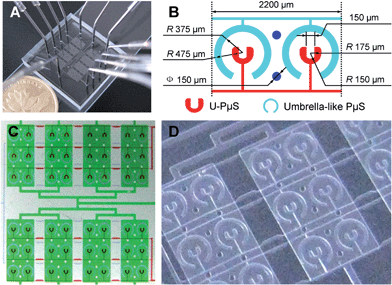
- (A)实际的整合PμS捕获装置。装置的入口和出口(直径:0.35 mm)由不锈钢管(外径0.5 mm)连接到聚乙烯管(内径:0.42 mm;长度:12 cm)。
- (B)工作区域的设计。U形PμS(宽:75-100 μm;面积:0.054 mm2)的非均匀平面构造在此指定为特定的三维尺寸。一一对应于U-PμSs的伞样PμS(宽:100 μm;面积0.23 mm2)被特地设计用于定量保护捕获细胞。
- (C)实际的PμS阵列捕获芯片的光学显微镜图。各种通道被装载入食用色素以可视化装置不同部分:绿色为流体通道,红色为U-PμSs,浅青绿为伞样PμSs。
- (D)实际的PμSs。
Fig.3 Fourfundamental states ofthe U-PμS
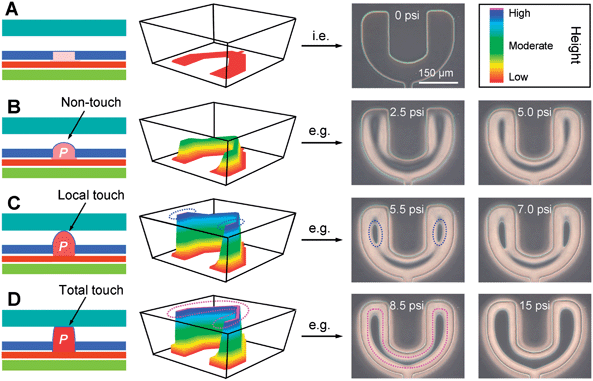
- (A)休眠模式。U-PμS在0 psi气动驱动下显示为二维结构。
- (B)初始动作模式。U-PμS显示了三维结构但没有接触到微腔。U-PμS和微腔天花板的间距能使细胞通过。
- (C)半动作模式。两边的U-PμS接触到了天花板,用两个椭圆虚线(蓝色)标记。在U-PμS的后方仍然有小间隙。这个现象是因为图2B所展示的U-PμS的非均匀的平面构造。
- (D)完全动作模式。U形U-PμS与微腔的天花板完全接触,用圆弧虚线(粉色)标记。不同气动驱动(0、2.5、5、5.5、7、8.5和15 psi)下U-PμS实际的顶部视图,从概念上讲对应于不同的U-PμS状态。
Fig. 4 Investigation of the U-PμS-based cell trapping in the microfluidic device
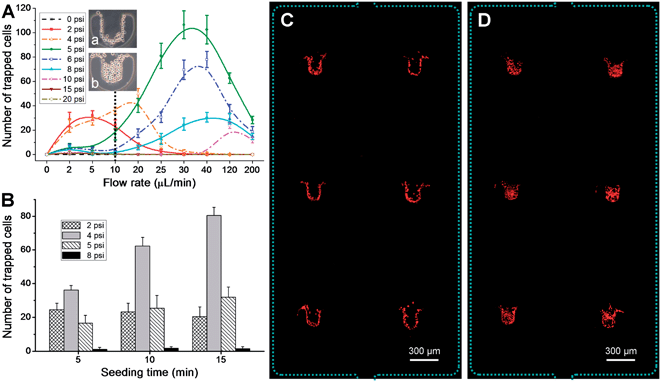
- (A)不同流速和气动驱动下,人肺腺癌A549细胞的捕获动力学。接种时间为5 min。结果显示,U-PμS可以提供不同数量的细胞捕获。线图显示细胞的U形排列(a),此时U-PμS的参数为气压4 psi以及流速20 μL min-1;细胞的完全排列(b),此时U-PμS的参数为气压5 psi以及流速30 μL min-1。
- (B)不同接种时间和气动驱动下,A549细胞的捕获动力学。流速为10 μL min-1(A中虚线)。结果显示,使用4 psi U-PμS捕获的细胞数量随着接种和捕获时间的增加而增加。然而使用2 psi U-PμS相对不变由于细胞丢失。
- (C)在气压4 psi以及流速20 μL min-1的条件下,平行捕获的A549细胞的荧光图像(PI核染色)。
- (D)在气压5 psi以及流速30 μL min-1的条件下,平行捕获的A549细胞的荧光图像。
Fig. 5 Comparison between the on (30 psi) and off (0 psi) conditions of the umbrella-like PμSs for the protection of trapped cells
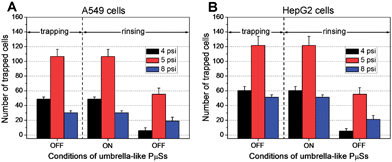
开(30 psi)和关(0 psi)条件下,进行伞样U-PμSs在120 μL min-1流速的冲洗中对(A)A549和(B)HepG2细胞的保护作用比较。在装置中,8个设计的腔室是对称的,这进一步导致平均分流进入腔室。本次测试中,U-PμSs捕获条件为:4 psi 20 μL min-1、5 psi 30 μL min-1和8 psi 40 μL min-1。在捕获过程中,检测伞样U-PμSs的在冲洗过程中的保护作用。结果显示,当伞样U-PμSs开启时,可以完全将细胞保留在U形U-PμSs中。然而当伞样U-PμSs关闭时,高流速条件下部分细胞被冲走。这个结果表明伞样U-PμSs在当前研究下可以很大程度解决捕获细胞丢失的问题,有效完成细胞保护。
Fig. 6 Three cell types at the same trapping condition

基于U-PμSs的捕获(A)人肝细胞癌HepG2细胞和(B)人乳腺癌MCF-7细胞。小图显示实际捕获(a)HepG2细胞在5 psi 25 μL min-1条件;(b)MCF-7在在5 psi 25 μL min-1条件。这个结果显示了U-PμSs捕获HepG2和MCF-7细胞同A549细胞具有相同的趋势(图4A)。
C图为不同流速和气动参数下,U-PμS中被捕获细胞的细胞活性。短时间(5 min)的接种和U-PμS捕获对细胞损害更小。小图显示了吖啶橙(acridine orange)和碘化丙啶(propidium iodide)染色的被捕获A549细胞的荧光染色图(4 psi 10 μL min-1)。
Conclusion
提出了一种气动驱动的在整合了U-PμS的芯片上的细胞捕获方法。
- 简单方便地实现基于精确控制的尺寸动力学,可逆、高通量、多数量的细胞捕获。
- 微结构可以出色的实现被捕获细胞的保护。
- U-PμSs的开/关非常适合基于培养的细胞分析,而不会受到固体微结构的物理干扰。
- 开关同样适合于动态捕捉和释放。
- U-PμSs可以实现阵列样细胞图案化和定位。
Reference
Liu W, Li L, Wang J, et al. Dynamic trapping and high-throughput patterning of cells using pneumatic microstructures in an integrated microfluidic device[J]. Lab on a Chip, 2012, 12(9): 1702–1709.